from chaos to beauty of code
Work with pandas.DataFrame can turn into an awkward pile of old (not so) good spaghetti code. My colleagues and I use this library often, and although we try to adhere to good programming practices such as modularizing code and unit testing, sometimes we still get in each other's way, creating confusing code.
I've put together some tips and pitfalls to avoid when making code in pandas clean. I hope they will be useful to you too. I will also refer to Robert Martin's classic book, Clean Code: Creating, Analyzing, and Refactoring.
TL;DR:
There is no one right way to write code, but here are some tips for working with pandas:
“No”
don't change
DataFrametoo much inside functions, because this way you can lose control over what and where will be added/removed from it;don't write methods that change
DataFrameand they don't return it because it's confusing.
“Yes”
Create new objects instead of changing the original one
DataFrameand don't forget to make a deep copy when necessary;perform only operations of a similar level within one function;
Design features with reusability in mind;
test your functions because it will help you create cleaner code, protect against bugs and edge cases, and document it.
Do not do it this way
First, let's look at a few flawed patterns inspired by real life. Later we will try to improve this code in terms of readability and control over what is happening.
Mutability
The most important thing to remember here is: pandas.DataFrame – This changeable objects [2, 3]. When you change a mutable object, it affects the same instance that you originally created, and its physical location in memory remains unchanged. In contrast, when you modify an immutable object (such as a string), Python creates a new object in a new memory location and changes the reference to the new object.
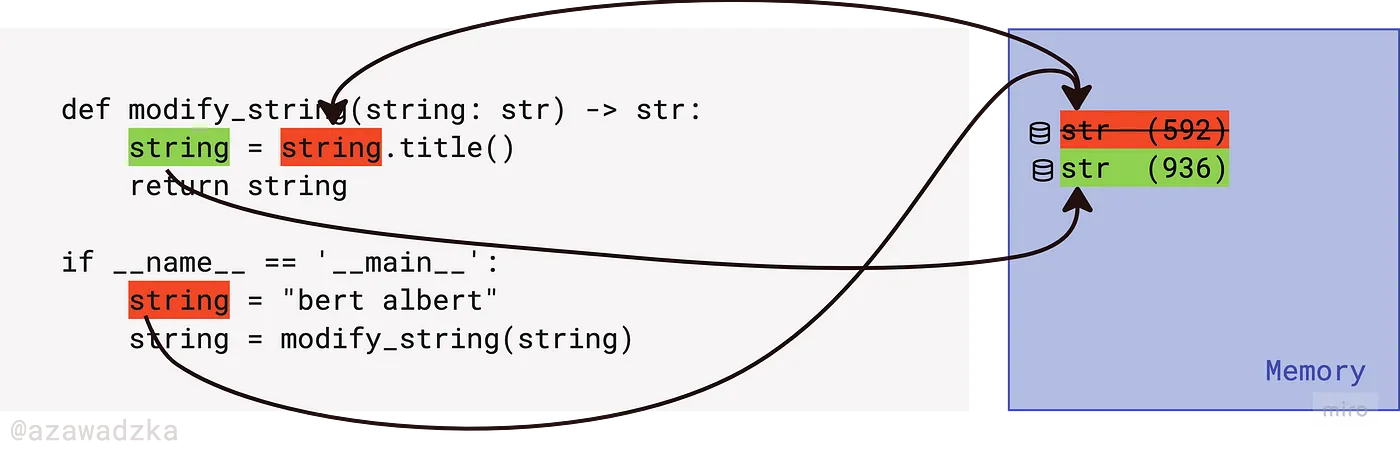
This is a very important point: in Python, objects are passed to a function by assignment [4, 5]. Look at the picture below: meaning df was assigned to the variable in_dfwhen it was passed to the function as an argument. And the original value dfAnd in_df inside functions point to the same memory location (the numeric value in parentheses), even if they have different variable names. During attribute modification, the location of the object being modified remains unchanged.

In fact, since we changed the original instance, return DataFrame and assigning it to a variable is redundant. This code gives exactly the same effect:

Note: the function now returns Noneso be careful not to overwrite df on Noneif you do the assignment: df = modify_df(df).
In contrast, if an object is immutable, it will change location in memory during modification, as in the example below. In the picture below, since the red string cannot be modified (strings are immutable), the green string is created as a new object occupying a new memory location. The string returned by the method is not the same string, whereas in the case of DataFrame the returned object would be exactly the same DataFrame.
The point is that changeDataFrame within functions has a global effect. If you don't remember this, you may:
accidentally change or delete part of the data, thinking that the action occurs only within the scope of the function, but this is not the case;
lose control over what is added and when
DataFramefor example, when calling nested functions.
Output arguments
We'll fix this problem later, but for now, one more no before we get to the yes.
The construct from the previous section is actually an antipattern called output argument [1 стр.45]. Typically, a function's input is used to produce an output value. If the only point of passing an argument to a function is to modify it, that is, the input argument changes its state, then this flies in the face of our intuition. This behavior is called a side effect. side effect) [1 стр.44] functions, and it should be well documented, or better yet, kept to a minimum, since it forces the programmer to remember what is happening in the background, and therefore increases the likelihood of making mistakes.
When we read a function, we are used to the idea of information going in to the function through arguments and out through the return value. We don't usually expect information to be going out through the arguments. [1 p.41]
When we read a function, we are accustomed to the fact that information enters the function through arguments and comes out through the return value. We usually don't expect information to be returned through arguments. [1 p.41]
The situation gets even worse if the function has dual responsibility: it both modifies the input and returns the output. Consider this function:
def find_max_name_length(df: pd.DataFrame) -> int:
df["name_len"] = df["name"].str.len() # side effect
return max(df["name_len"])As you would expect, it returns a value, but it constantly modifies the original DataFrame. The side effect takes you by surprise – nothing in the function signature indicated that our input would be affected. In the next step we will look at how to avoid this anti-pattern.
It should be like this!
Minimizing object modifications
To eliminate the side effect, in the code below we have created a new temporary variable instead of modifying the original one DataFrame. Designation lengths: pd.Series indicates the data type of the variable.
def find_max_name_length(df: pd.DataFrame) -> int:
lengths: pd.Series = df["name"].str.len()
return max(lengths)This function design is better because it encapsulates an intermediate state rather than creating a side effect.
Another warning: please be aware of the differences between deep and shallow copying [6] elements from DataFrame. In the above example we have changed every element of the original series df["name"]so old DataFrame and the new variable do not have any common elements. However, if you directly assign one of the original columns to a new variable, the underlying elements will still have the same references in memory. Here are examples:
df = pd.DataFrame({"name": ["bert", "albert"]})
series = df["name"] # поверхностная копия
series[0] = "roberta" # <-- изначальный DataFrame изменяется
series = df["name"].copy(deep=True)
series[0] = "roberta" # <-- изначальный DataFrame не изменяется
series = df["name"].str.title() # в любом случае не копия
series[0] = "roberta" # <-- изначальный DataFrame не изменяетсяYou can withdraw DataFrame after each step to monitor what is happening. Remember that when creating a deep copy, new memory will be allocated, so it is worth considering whether you need to save memory in your case.
Grouping similar operations
Perhaps for some reason you want to save the length calculation result. Still shouldn't add it to DataFrame inside a function due to possible side effects, and also due to the accumulation of multiple responsibilities in a single function.
I like the rule One Level of Abstraction per Functionwhich reads:
We need to make sure that the statements within our function are all at the same level of abstraction.
Mixing levels of abstraction within a function is always confusing. Readers may not be able to tell whether a particular expression is an essential concept or a detail. [1 p.36]
We need to make sure that all actions within our function are at the same level of abstraction.
Mixing levels of abstraction in a function always leads to confusion. The reader may not understand whether a particular expression is an essential concept or a detail. [1 стр.36]
Let's also use the principle of sole responsibility [1 стр.138] from OOP, although we are not currently focusing on object-oriented code. (And in principle, OOP, even in the context of Python, is the last thing you associate with data analysis using pandas. (note by the author of the translation))
Why not prepare the data in advance? Let's divide the data preparation and the actual calculations into separate functions:
def create_name_len_col(series: pd.Series) -> pd.Series:
return series.str.len()
def find_max_element(collection: Collection) -> int:
return max(collection) if len(collection) else 0
df = pd.DataFrame({"name": ["bert", "albert"]})
df["name_len"] = create_name_len_col(df.name)
max_name_len = find_max_element(df.name_len)Separate column creation task name_len was passed to another function. It does not change the original DataFrame and performs one task at a time. Later we will get the maximum element by passing the new column to another special function.
Let's tidy up the code using the following steps:
We can use the function
concatand move it to a separate functionprepare_datawhich would group all data preparation steps in one place;We can also use the method
applyand work with individual texts rather than series of texts;Let's not forget about the use of shallow and deep copying, depending on whether it is necessary or not to change the source data:
def compute_length(word: str) -> int:
return len(word)
def prepare_data(df: pd.DataFrame) -> pd.DataFrame:
return pd.concat([
df.copy(deep=True), # deep copy
df.name.apply(compute_length).rename("name_len"),
...
], axis=1)Let's reuse the code
The way we've divided the code makes it easy to come back to the script later and take the entire function and reuse it in another script, which is great!
There's one more thing you can do to increase reusability: pass column names as parameters to functions. There's a lot of refactoring out there, but sometimes flexibility and reusability come at a price.
def create_name_len_col(df: pd.DataFrame, orig_col: str, target_col: str) -> pd.Series:
return df[orig_col].str.len().rename(target_col)
name_label, name_len_label = "name", "name_len"
pd.concat([
df,
create_name_len_col(df, name_label, name_len_label)
], axis=1)Making the code testable
Have you ever found out that your preprocessing was wrong after weeks of experimenting with a preprocessed dataset? No? Lucky. In fact, I've had to repeat an entire series of experiments because annotations didn't work, which could have been avoided if I'd only tested a couple of basic features.
So, important scripts must be tested [1, с. 121, 7]. Even if the script is just a helper, I now try to test at least the most important, low-level functions. Let's go back to the steps we took from the beginning:
A bunch of different functions are tested here: calculating the length of a name and aggregating the result for an element
max. And the test failsAttributeError: Can only use .str accessor with string values!although we didn't expect it, did we?
def find_max_name_length(df: pd.DataFrame) -> int:
df["name_len"] = df["name"].str.len() # побочный эффект
return max(df["name_len"])
@pytest.mark.parametrize("df, result", [
(pd.DataFrame({"name": []}), 0), # упс, здесь тест упадет
(pd.DataFrame({"name": ["bert"]}), 4),
(pd.DataFrame({"name": ["bert", "roberta"]}), 7),
])
def test_find_max_name_length(df: pd.DataFrame, result: int):
assert find_max_name_length(df) == resultThis is much better – we focused on one task, so the test became easier. Additionally, we don't have to get stuck on column names like we did before. However, I still feel that the data format interferes with checking the correctness of the calculations.
def create_name_len_col(series: pd.Series) -> pd.Series:
return series.str.len()
@pytest.mark.parametrize("series1, series2", [
(pd.Series([]), pd.Series([])),
(pd.Series(["bert"]), pd.Series([4])),
(pd.Series(["bert", "roberta"]), pd.Series([4, 7]))
])
def test_create_name_len_col(series1: pd.Series, series2: pd.Series):
pd.testing.assert_series_equal(create_name_len_col(series1), series2, check_dtype=False)Here we have put things in order and are testing the computational function itself without a wrapper in the form
pandas. It's easier to come up with edge cases if you focus on one thing at a time. I realized that I want to check the valuesNonewhich may appear inDataFrame, and I ended up having to improve my function to make the test pass. Bug caught!
def compute_length(word: Optional[str]) -> int:
return len(word) if word else 0
@pytest.mark.parametrize("word, length", [
("", 0),
("bert", 4),
(None, 0)
])
def test_compute_length(word: str, length: int):
assert compute_length(word) == lengthWe only need a test for
find_max_element:
def find_max_element(collection: Collection) -> int:
return max(collection) if len(collection) else 0
@pytest.mark.parametrize("collection, result", [
([], 0),
([4], 4),
([4, 7], 7),
(pd.Series([4, 7]), 7),
])
def test_find_max_element(collection: Collection, result: int):
assert find_max_element(collection) == resultAnother benefit of unit testing that I never forget to mention is that it is a way of documenting your code as someone who doesn't know what's going on there (for example, you are from the future)can easily determine the inputs and expected outputs, including edge cases, just by looking at the tests.
Conclusion
These are a few techniques that I have found useful when writing code and reading other people's code. I'm not saying that one way or another is the right way to code – it's up to you to decide whether you want a fast experience or a polished and tested codebase. But I hope this article will help you structure your scripts so that they are prettier and more reliable.
I will be glad to receive feedback and other comments. Happy coding!
List of sources
The illustrations were created using Miro.