“Look, this is something”: self-replication of artificial DNA

One of the main distinguishing features of any living organism is the ability to preserve and reproduce the necessary information to create their own kind. This primarily manifests itself in DNA replication, when a pair of daughter ones appear from the same parent DNA, which are an exact copy of their progenitor. In nature, this process is observed everywhere, however, it is extremely difficult to recreate it in the laboratory from scratch, however, it is quite realistic.
Scientists from the Institute of Biochemistry. Max Planck (Germany) successfully created a biological system that has the ability to replicate its own DNA. What techniques were used to create synthetic replicating DNA, how effective is the resulting system, and what does this discovery mean for modern synthetic biology? We will find answers to these questions in the report of scientists. Go.
Study basis
In the field of creating artificial biological systems, the ability to replicate is a key aspect of the usefulness of the created system. To achieve this, according to scientists, it is possible through the implementation of the basic components: replication *, transcription* and broadcast * DNA
DNA replication * – The process of creating two daughter DNA molecules from one maternal DNA.
Transcription* – the process of RNA synthesis using DNA as a template, i.e. The process of transferring genetic information from DNA to RNA.
Broadcast * – The process of protein synthesis from amino acids on an RNA matrix carried out by a ribosome.
In other words, in order to create a full-fledged artificial life, it is necessary to implement self-encoded reproduction, i.e. self-replication. This is an extremely complicated and confusing process.
The authors of the study note that in systems based on existing biochemistry, for example, MPC (minimal protein-based cells)Self-replication requires a complete rethinking of molecular biology, including DNA replication, transcription and translation, taking into account a completely cell-free environment.
The synthesis of protein from DNA can be achieved in certain recombinant systems based on phage * RNA polymerases are the main parts of the translation mechanism of Escherichia coli and the system of minimal energy recovery, i.e. PURE – Protein synthesis Using Recombinant Elements (protein synthesis using recombinant elements).
Bacteriophages or phages * – viruses that selectively infect bacterial and archaeal cells. In biology it is used as a vector (DNA for transferring genetic material into the cell).
Meanwhile, it remains extremely difficult to recreate the process of DNA replication of the genome based on transcription and translation (TTcDR – transcription – translation-coupled DNA replication) encoding all macromolecular components of the PURE system using self-encoded replicas *.
Replicom * – multi-protein complex, due to which the replication of DNA of the bacteria takes place.
DNA replication based on DNA polymerases (DNAP) from phages (e.g., Phi29) can help implement the TTcDR technique.
However, in the TTcDR method, the full-sized Phi29 genome could be achieved only by blocking some of the replicating factors.
All the above methods have their theoretical advantages, but in practice they did not give a full result. In the work we are considering today, scientists describe a translation system that provides self-encoded replication and expression of large DNA genomes under well-defined cell-free conditions. In particular, self-replication of the genome of over 116 kb was achieved. (thousand pairs of nucleotides), covering the full range of translation factors Escherichia coli (Escherichia coli), all three ribosomal RNAs, an energy regeneration system, as well as RNA- and DNA-polymerase *.
Polymerase * – an enzyme that performs the synthesis of polymers of nucleic acids. DNA polymerase synthesizes DNA, and RNA polymerase synthesizes RNA. This process proceeds through complementary copying of parental DNA or RNA.
In addition, the created system demonstrated the ability to synthesize more than 30 translation factors, half of which expresses in amounts equal to or greater than the introductory ones.
Research results
At the first stage, scientists tested the self-encoded Phi29-DNAP-dependent TTcDR using the standard system protocol PURExpress.
Phi29-DNAP coding region flanked promoter * T7, first cloned into the pCR-Blunt TOPO vector (pREP, 1a)
Promoter * Is the DNA nucleotide sequence used by RNA polymerase as a region for the start of transcription.
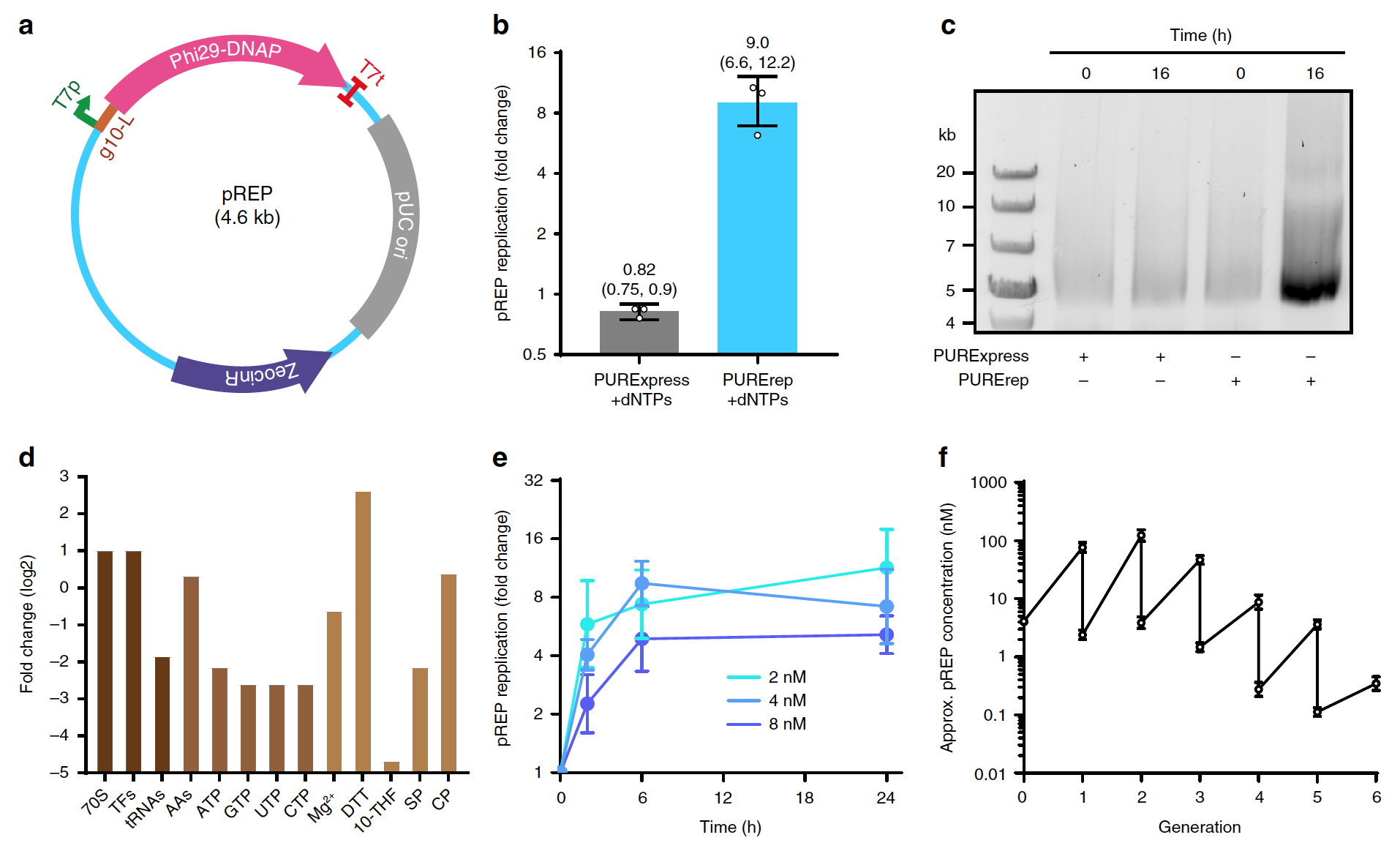
Image No. 1
In theory, this architecture should provide spontaneous RNA rolling ring replication * using self-encoded DNAP without additional replication proteins or externally provided DNA primers.
Rolling Ring Type Replication * – the process of unidirectional nucleic acid replication, during which there is a rapid synthesis of multiple copies of ring DNA or RNA molecules.
However, using the standard PURExpress reaction with dNTP (nucleotide containing deoxyribose C5H10O4) and 4 nM pREP (LB medium supplemented with zeocin C55H86N20O21S2), it was not possible to detect DNA synthesis using agarose gel electrophoresis *nor with real-time polymerase chain reaction * (1b and 1c)
Agarose gel DNA electrophoresis * – a method for separating DNA fragments along their length, based on determining the speed of movement of fragments of different lengths at the moment of movement in the gel under the influence of an external electric field.
Real-time polymerase chain reaction * – a method for simultaneously creating additional copies of DNA sections and measuring the amount of a given DNA molecule.
To improve DNA replication without the involvement of specialized PURE systems, it was decided to optimize the standard PURExpress reaction protocol. For this, the relative number of translation factors, ribosomes, and a reducing agent was increased while reducing the levels of tRNA (transport RNA) and rNTP (ribonucleoside triphosphate) (1d)
Using this optimized PURE composition (PURErep), it was possible to achieve ∼5–12-fold replication of pREP monomeric units in TTcDR reactions (1b and 1e) Full-size pREP synthesis was confirmed MluI cleavage * replication product (1c)
MluI * is a commercially available restriction endonuclease, i.e. an enzyme that catalyzes the hydrolysis of nucleic acids.
The expression of green fluorescent protein (sfGFP) was used as an evaluation factor of translation activity. It was found that a change in the composition of PURE led to a decrease in protein synthesis by 20-40% compared with PURE without TTcDR. However, this decrease in protein expression remains acceptable given the improved compatibility of the PURErep system with DNA replication.
QPCR-based DNA replication analysis revealed a stable doubling time (1-2 hours) for various initial matrix concentrations, while DNA replication continued even after 24 hours at 30 ° C (1st)
TTcDR was also stable for more than five generations of sequential dilution when only 4% of the finished PURErep / pREP reaction product was directly transferred to the new PURErep mixture (1f) The main conclusion of this observation is that TTcDR products can serve as templates for DNA self-replication over several generations.
Scientists note that a significant amount of product with low electrophoretic mobility is typical for replication as a rolling ring, which was observed during the experiments. These reaction products may be presented. concatemers * high molecular weight and / or DNA / MgPPi clusters.
Concatemer * – multiple copies of DNA sequences assembled in a sequential cluster.
The formation of products with a size of ~ 5 kb was also found in untreated samples. It follows that TTcDR reactions can produce significant amounts of pREP monomeric copies.
In addition, scientists were able to transform products E.coli after removal of the parent plasma. The resulting purified reaction products were identical in size to the monomeric pREPs.
At the next stage, scientists decided to create a set of genes encoding important components of the PURE reaction: 31 essential translation factor (FT) of the bacterium E. coli.
To do this, a study of TTcDR from pREP (4.6 kb) was performed in conjunction with each of the three large plasmids *: pLD1 (30 bp, 13 FT), pLD2 (20 bp, 8 FT) and pLD3 (23 bp, 9 FT). All were cloned to provide recombinant expression of 30 of the 31 bacterial translation factors. E. coli.
Plasmids * – small DNA molecules capable of self-replication.
TTcDR products of all four plasmids (including pREP) showed identical MluI restriction features: clonal plasmids, usually propagated in E.coli (2a)
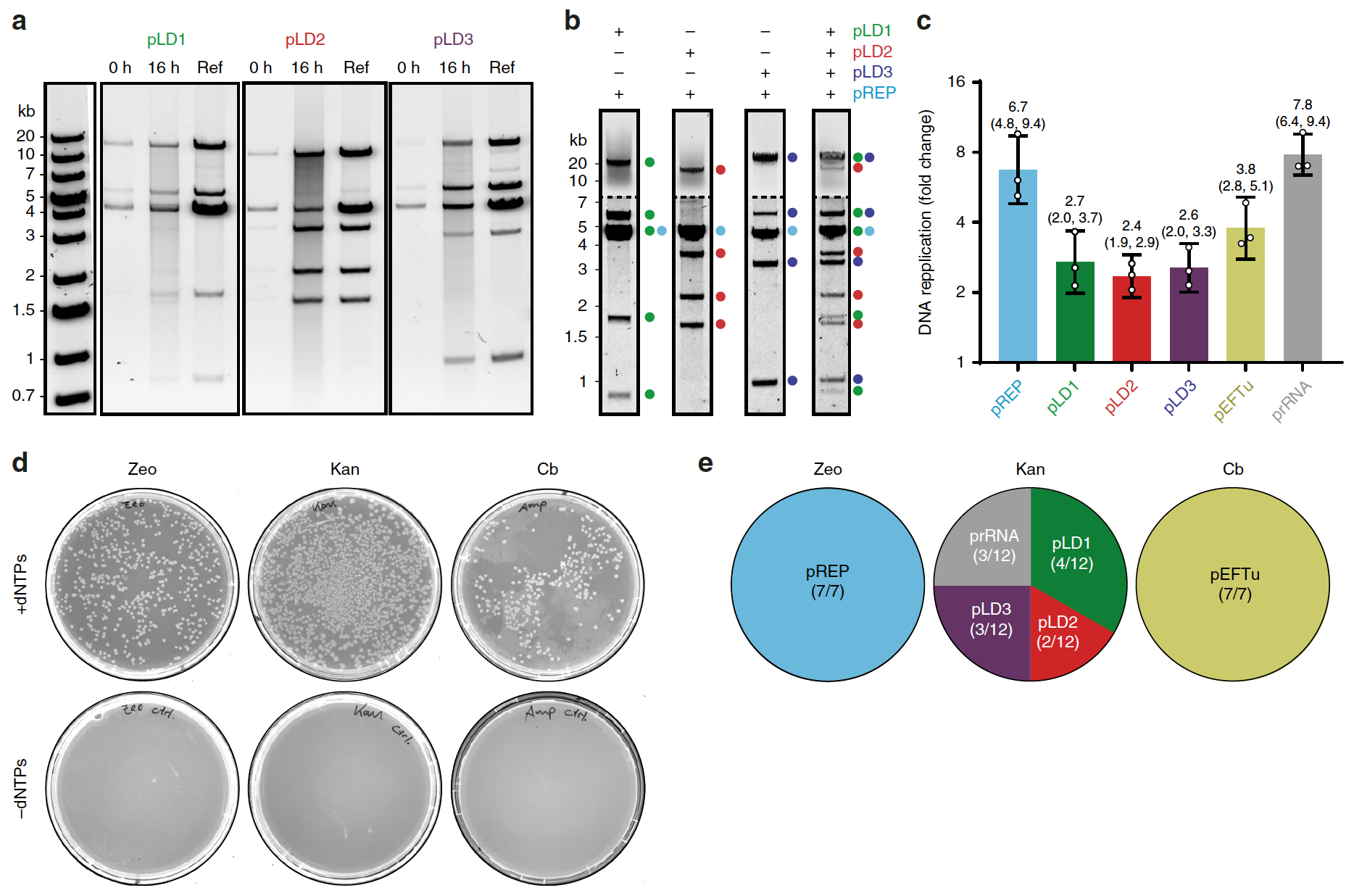
Image No. 2
It is also worth noting that pLD TTcDR products can be directly transformed into E.coliwhere they are stored as monomeric plasmids.
The modified PURErep mixture also provided complete replication of all three pLD plasmids together with PURErep during the reaction in a common medium (2b)
Scientists did not stop there and already at the next stage of the study they tried to expand the genetic load of the TTcDR system by co-replicating plasmids encoding additional components of the PURE system: EF-Tu (pEFTu), which is absent in the pLD system, as well as ribosomal RNA-operon * rrnB (prRNA precursor rRNA), which encodes 23S rRNA (ribosomal RNA), 16S rRNA, 6S rRNA and tRNA (transport RNA, in this case Glu2) (2c)
Operon * – a functional unit of the unicellular genome, which includes genes encoding proteins.
QPCR experiments targeting plasmid-specific amplicons *, confirmed that the monomeric units of all six plasmids (total DNA length was 93 kb) replicated approximately 2-8 times compared to the initial number (2c)
Amplicon * – extrachromosomal amplification unit (formation of copies of DNA sections).
Additional confirmation of the success of joint replication of plasmids was the formation of colonies resistant to either zeocin (pREP), or to kanamycin (plasmid pLD and prRNA), or to carbenicillin (pEFTu). These colonies were the result of the transformation of PURErep reaction products treated with DpnI (2d)
The selected 26 clonal colonies followed by analysis of restriction features once again confirmed the success of TTcDR of all six plasmids (2e)
Using the same method as previously described, scientists conducted another procedure for the joint replication of five additional plasmids: genes for the minimal regeneration system of nucleoside triphosphate based on creatine kinase (pCKM), adenylate kinase (pAK1) and nucleoside diphosphate kinase (pNDK); as well as T7 RNA polymerase (T7RNAP) and pyrophosphatase (pIPP).
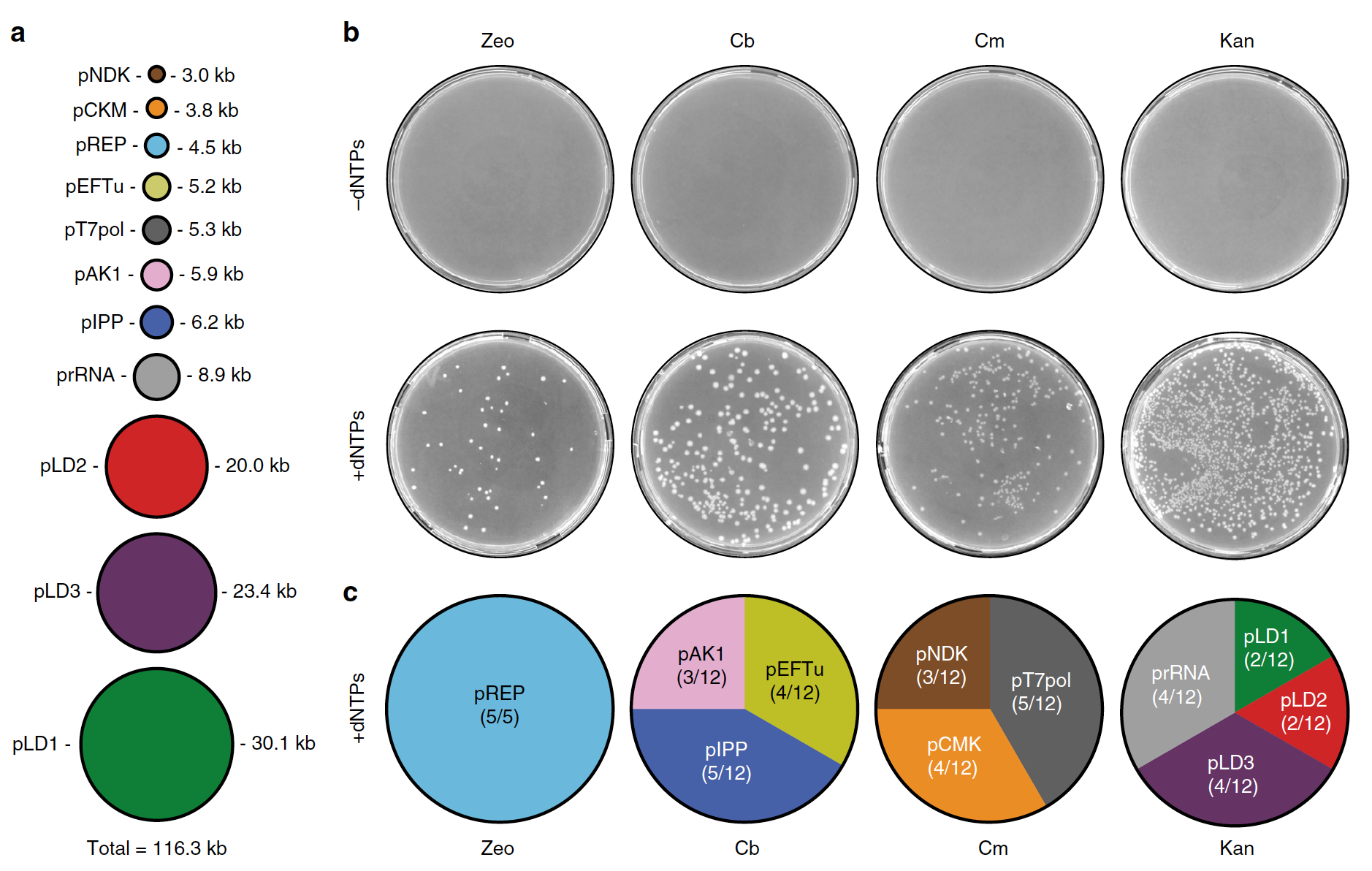
Image No. 3
With a total size of 116.3 kb this set of 11 plasmids reaches> 100% of the estimated genome length proposed for a minimal, self-replicating system that depends only on low molecular weight nutrients (3a)
The scientists then decided to test whether PURErep can provide parallel gene expression during replication. For this, the multicistronic expression of FT encoded on three pLD plasmids: pLD1, pLD2 and plD3 (not including pEFTu) was examined.
In order to investigate whether PURErep is able to generally support multicistronic expression from these plasmids, cell-free expression was performed from each individual plasmid in the presence of BODIPY-Lys-tRNALys, which allows fluorescence labeling of translation products.
In order to improve detection sensitivity and enable the quantification of newly synthesized proteins, a quantitative analysis of protein expression based on mass spectrometry using stable isotopic labels was additionally carried out.
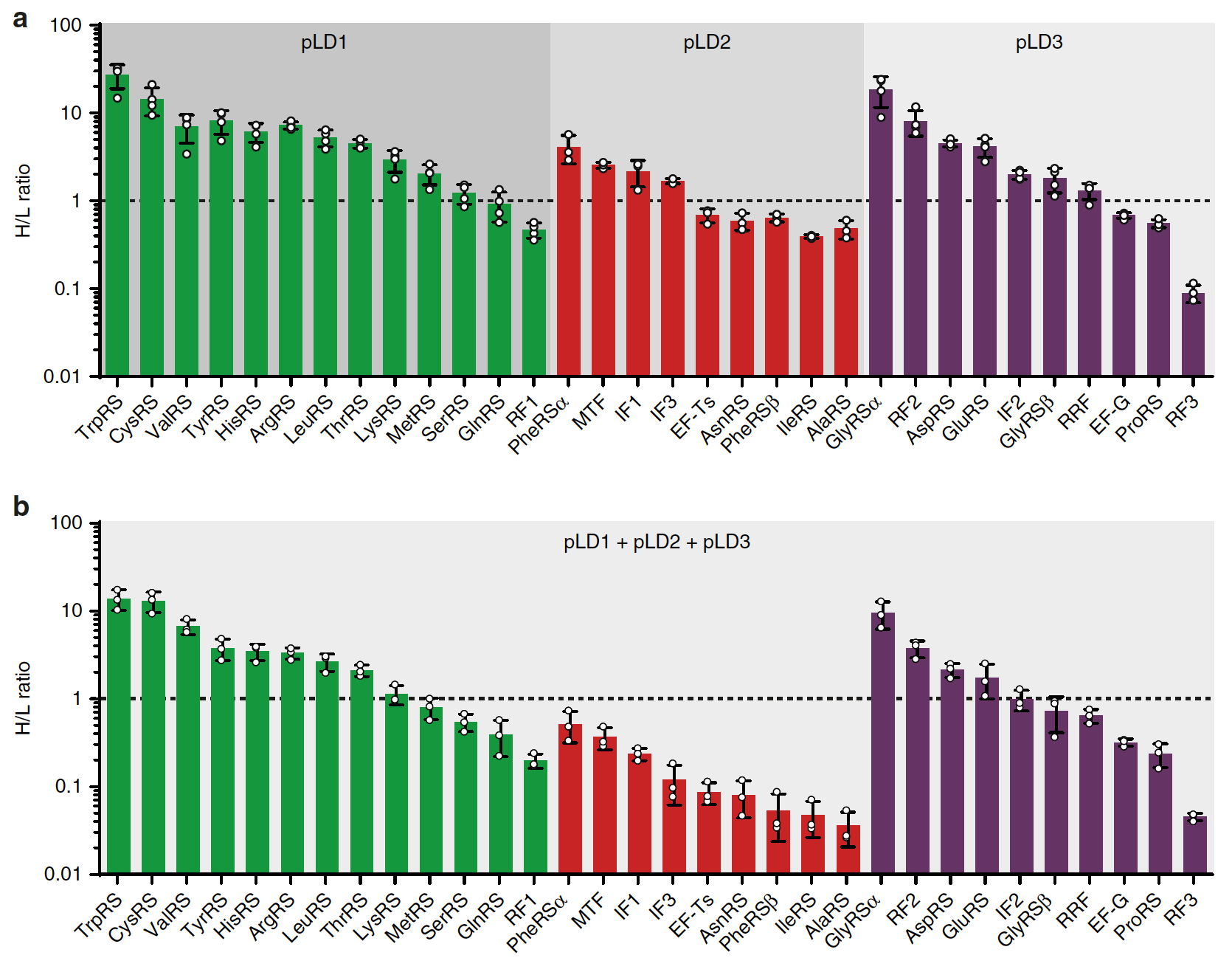
Image No. 4
This analysis provided strong evidence for the synthesis of all FT subunits of proteins encoded by pLD (4a) Partial or complete regeneration of proteins encoded in both pLD2 and pLD3 was also observed.
Analysis of expression showed that even in unmodified PURErep in combination with pREP, there is a complete replication of 32 FT cidrons encoded by pLD, and expression of about half of the encoded FT peptide chains, the number of which corresponds to or exceeds the initial one.
For a more detailed acquaintance with the nuances of the study, I recommend a look at report of scientists and Additional materials to him.
Epilogue
In this work, scientists managed to achieve what many of their colleagues could only dream of before – to create an artificial system that is capable of self-replicating.
Exaggerated saying, this means that they were able to take the first steps towards creating a system that will reproduce itself, thereby simulating biological processes as much as possible.
The authors of the study are extremely pleased with the results and plan to expand the artificial genome with additional DNA segments in the future. They want to create a system with a shell that can maintain viability by absorbing nutrients and disposing of waste. Such an artificial structure can be used as a specialized production bio-device for natural substances or as a basis for creating even more complex systems.
Cloning is a complex process, of which there is no doubt, but the creation of a complete, viable synthetic system is a completely different level. Someone will say that such studies are dangerous, because no one has the right to assume the responsibilities of mother nature. However, a person’s curiosity is almost impossible to calm down. Our desire to understand everything and repeat everything is one of the driving factors in the progress of science. For better or worse, this is a question for which there is no single answer. We can only admire the achievements of bright minds and cautiously look to the future, while hoping for a utopia.
Thank you for your attention, stay curious and have a great weekend everyone, guys! 🙂
A bit of advertising 🙂
Thank you for staying with us. Do you like our articles? Want to see more interesting materials? Support us by placing an order or recommending to your friends, cloud VPS for developers from $ 4.99, A unique analogue of entry-level servers that was invented by us for you: The whole truth about VPS (KVM) E5-2697 v3 (6 Cores) 10GB DDR4 480GB SSD 1Gbps from $ 19 or how to divide the server correctly? (options are available with RAID1 and RAID10, up to 24 cores and up to 40GB DDR4).
Dell R730xd 2 times cheaper at the Equinix Tier IV data center in Amsterdam? Only here 2 x Intel TetraDeca-Core Xeon 2x E5-2697v3 2.6GHz 14C 64GB DDR4 4x960GB SSD 1Gbps 100 TV from $ 199 in the Netherlands! Dell R420 – 2x E5-2430 2.2Ghz 6C 128GB DDR3 2x960GB SSD 1Gbps 100TB – from $ 99! Read about How to Build Infrastructure Bldg. class using Dell R730xd E5-2650 v4 servers costing 9,000 euros for a penny?



